Tool: There are following facilities are available under Tool Section:
1. RiPP Class Prediction Using Genomic Sequences
2. RiPP Precursor Prediction
3. RiPP Cleavage & Crosslinks Prediction
1. RiPP Class Prediction using Genomic Sequences
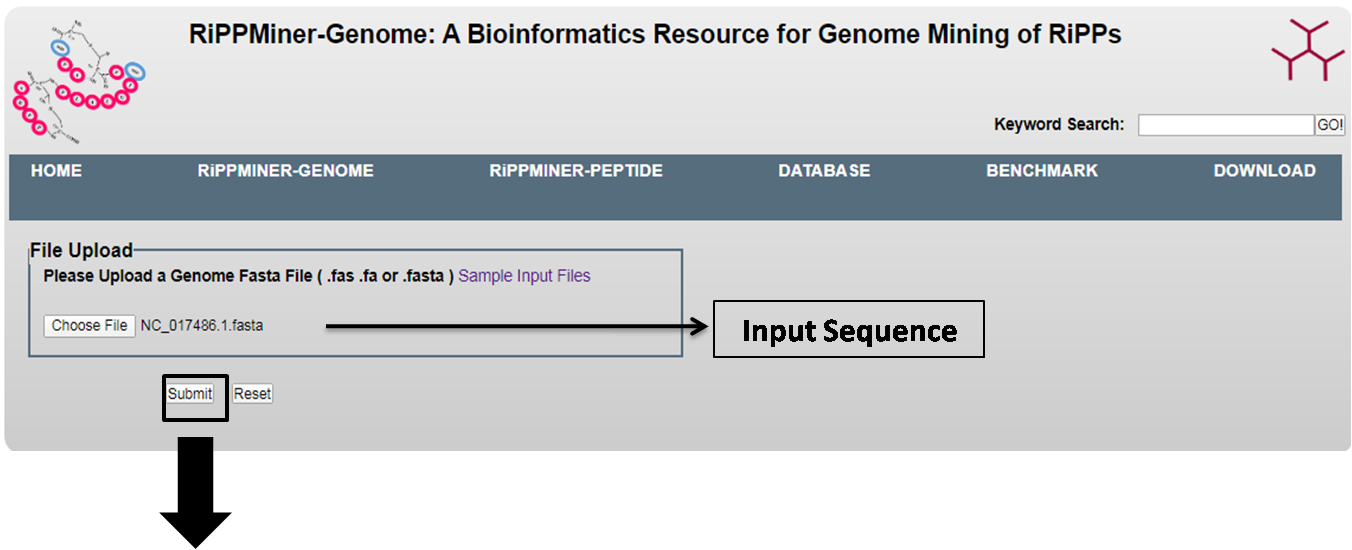
This RiPPMiner-Genome takes genomic sequences in fasta format as an input.
RiPPMiner-Genome can be use to predict nine classes of RiPP Clusters.:
These nine classes are :Head-to-tail cyclized peptide, Lanthipeptide, Cyanobactin, Lassopeptide, Thiopeptide, LAPs, Linaridin, Bottromycin,Glycocin.
It can also predict the three major subclasses of Lanthipeptides (i.e. Lanthipeptides A, B and C).
1.1. RiPP Cluster and Class Prediction
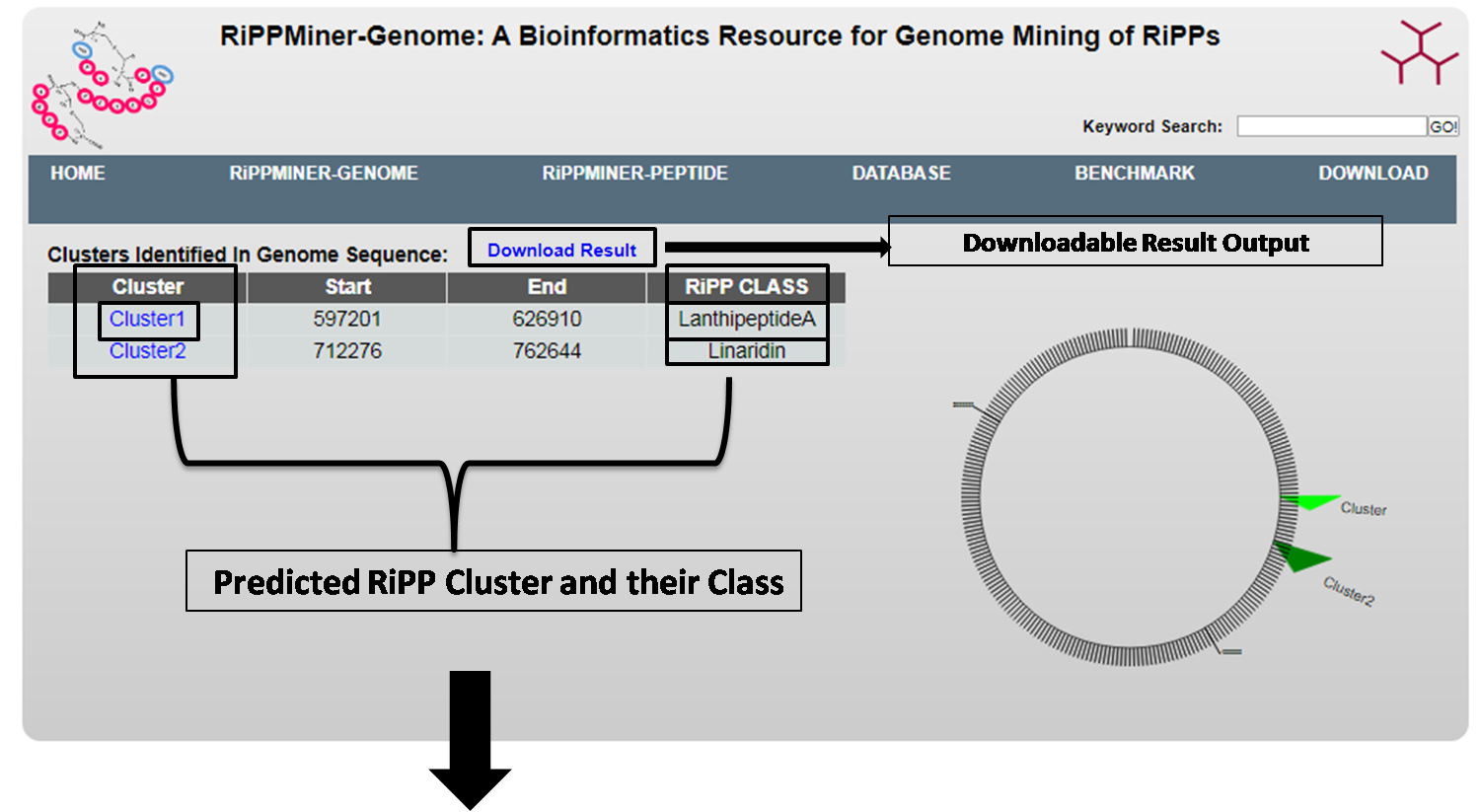
RiPPMiner-Genome predicts RiPP clusters, their start and end coordinates within the genomic region and which RiPP class it belongs.
2 & 3. RiPP Precursor,Cleavage and Cross-link Prediction
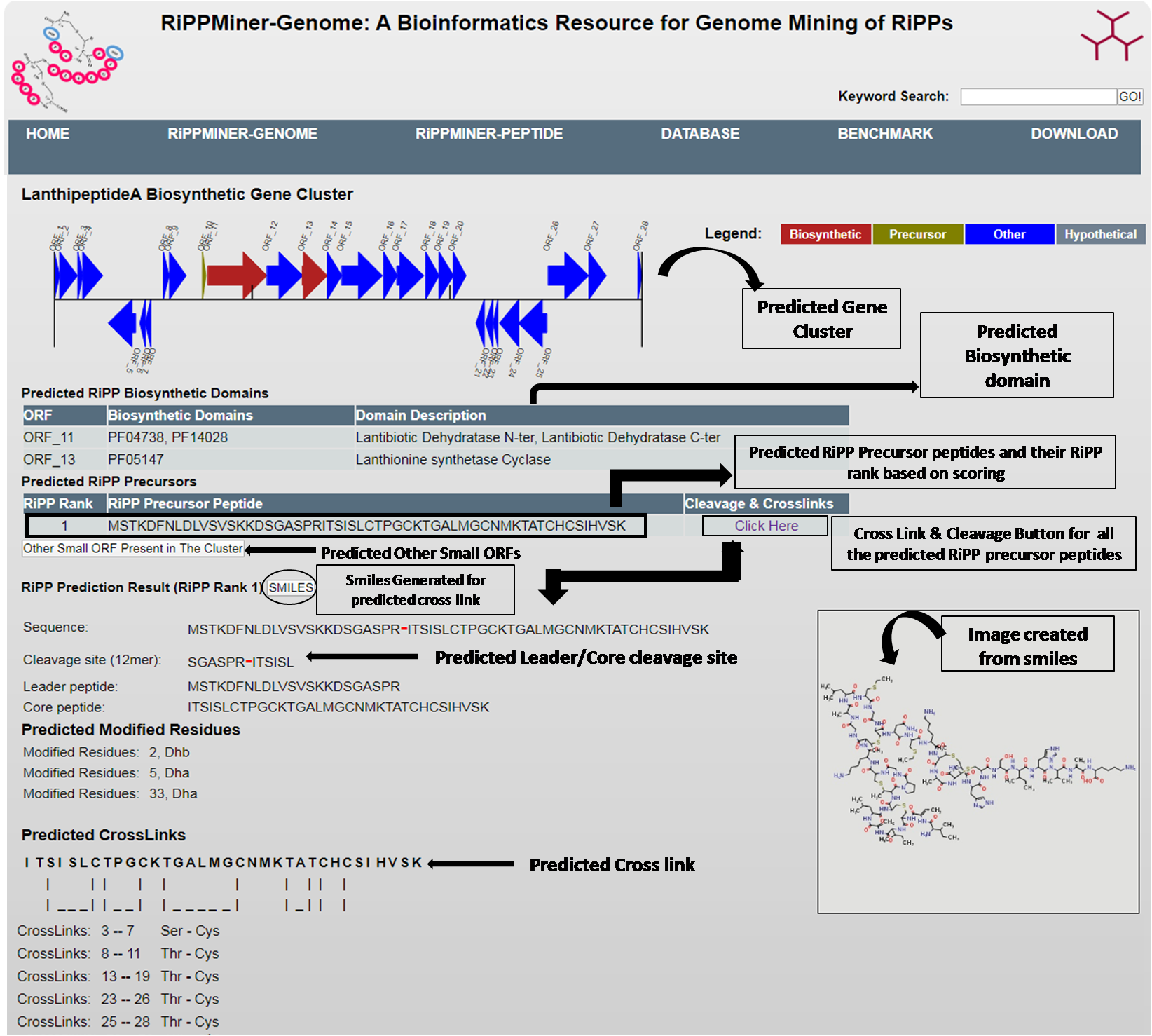
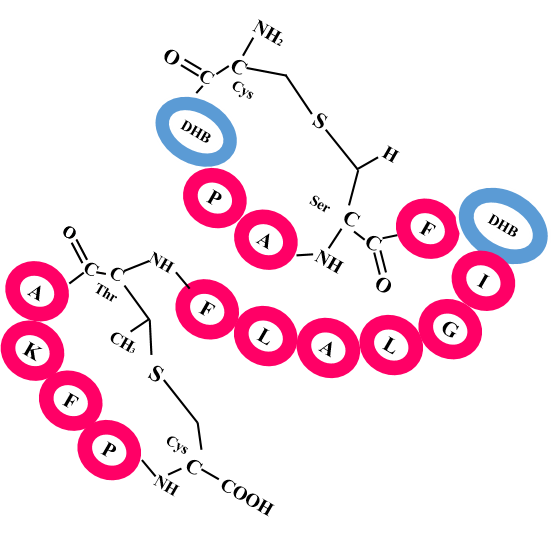
By clicking the each cluster link RiPPMiner- Genome will give the graphical cluster information,clusters biosynthetic domains information and their function.It will predict RiPP Precursor,their cleavage site with in the RiPP cluster using Classifier.
RiPPMIner-Genome also predicts cross links within the core peptide of four major RiPP Classes i.e.
LANTHIPEPTIDE
THIOPEPTIDE
LASSOPEPTIDE
CYANOBACTIN